Chemical and Biophysical Principles underlying cell-cell juxtacrine signaling
 Cell–cell communication by juxtacrine signaling plays a key role in many developmental, physiological, immunological, neural, and pathological processes. Signals exchanged between neighboring cells via Notch amplify cellular differences, tipping the scales of cell fate determination and enabling pattern formation during development. Juxtracrine signaling via neurexin-neuroligin directs synaptogenesis and synaptic remodeling. The juxtacrine interaction between T cell receptors and checkpoint receptors (e.g., CTLA-4, PD-1) with respective ligand proteins presenting on antigen presenting cells establishes an immunological synapse, regulating the immunological responses of cells.
Cell–cell communication by juxtacrine signaling plays a key role in many developmental, physiological, immunological, neural, and pathological processes. Signals exchanged between neighboring cells via Notch amplify cellular differences, tipping the scales of cell fate determination and enabling pattern formation during development. Juxtracrine signaling via neurexin-neuroligin directs synaptogenesis and synaptic remodeling. The juxtacrine interaction between T cell receptors and checkpoint receptors (e.g., CTLA-4, PD-1) with respective ligand proteins presenting on antigen presenting cells establishes an immunological synapse, regulating the immunological responses of cells.
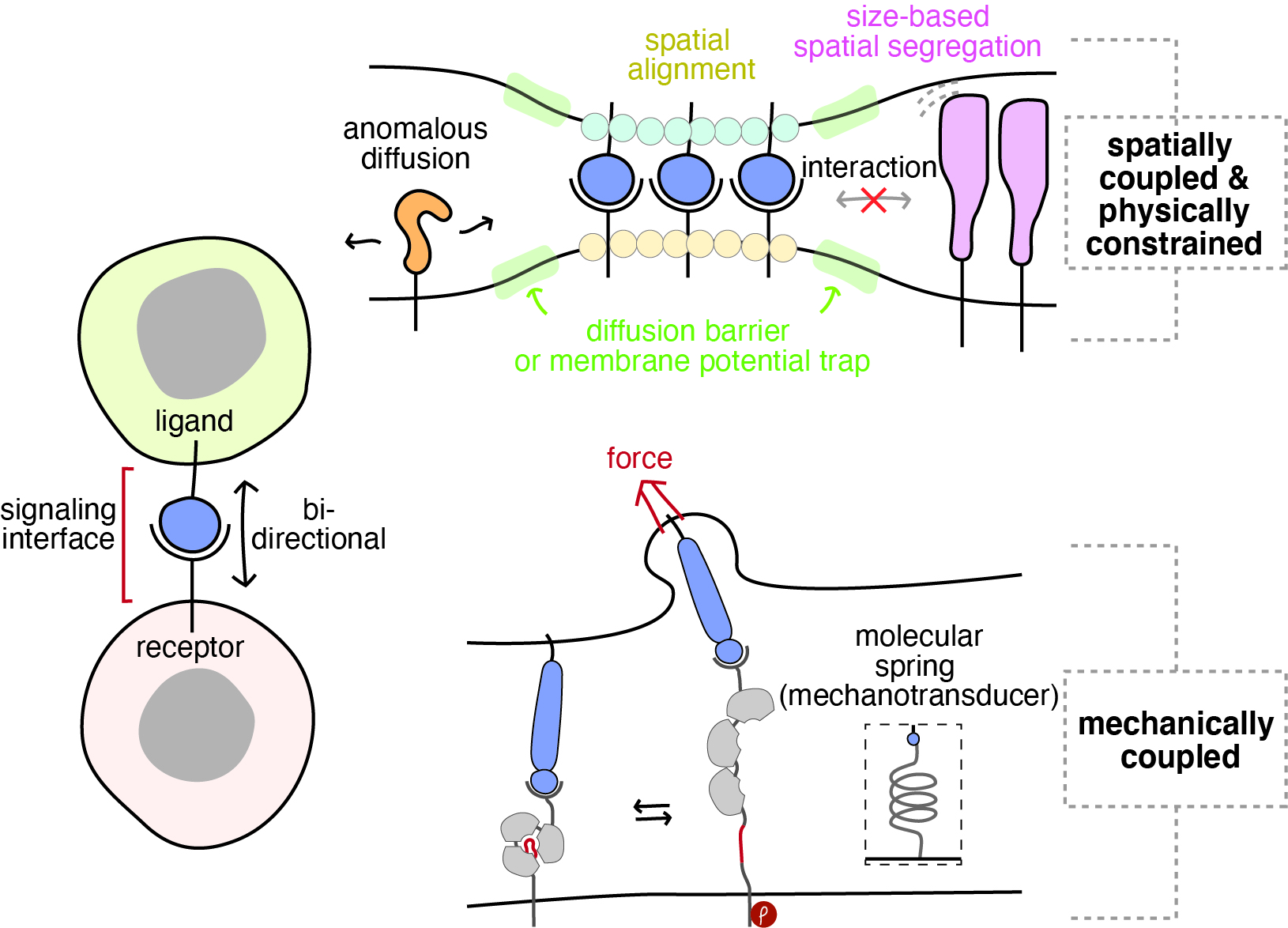
Due to the nature of signal transduction via physical contacts, the operating mechanisms and dynamics underlying juxtacrine signaling are quite different, compared with more traditional soluble ligand-mediated cell signaling (Figure). Signal exchange is localized to the signaling interface between the two neighboring cells and can be bidirectional and asymmetric. Additionally, the signaling surfaces undergo drastic reorganizations of the protein and lipid components as cell surfaces engage in juxtacrine ligand-receptor interaction. More importantly, through ligand-receptor engagement and surface reorganization, signaling complexes in the juxtaposed cells are coupled spatially and mechanically. These unique features allow cells sensing and responding to the complex and dynamic properties of the surroundings precisely, directing a wide range of cellular functions and orchestrating the development of multicellular organisms. Despite remarkable progress during past decades, our understanding on the juxtacrine signaling is mostly limited to genetic, structural, and biochemical information, and how the spatial and mechanical properties of the signaling interface orchestrate the dynamics of signaling components is still unclear. Since traditional genetic and molecular biology approaches cannot capture these physical dynamics alone, interrogation of the juxtacrine signaling dynamics has been challenging.
Develop cutting-edge nanotechnology tools for cell-cell signaling
To address this challenge, we use non-traditional chemical and biophysical approach. Particularly, as a highly interdisciplinary team led by a chemist and nanoscientist PI, our strategy is to develop enabling-nanotechnology tools that can sense and manipulate spatiotemporal and mechanical dynamics of juxtacrine receptors. We employ nanotechnology tools because of its unique capacity to integrate multiple functions into the tool, enabling the designer’s approach to add any desired components with unique functions, which do not exist in biology, into a single integrated system. These tools can provide an unprecedented means of measuring, perturbing, and analyzing cell signaling processes. As an initial example, we develop novel single-cell perturbation techniques (e.g., mechanogenetics) to manipulate spatial, physical (e.g., elimination of physical barriers), and mechanical properties of juxtacrine receptors and their environments systematically and quantitatively. Separately, we develop techniques that can sense physical and mechanical properties of cellular microenvironments (e.g., in vivo micropressure sensors). Diffusional and assembly dynamics of juxtacrine receptors regulate the signaling dynamics. Hence, we develop targeted and nonperturbing single-molecule nanoparticle probes that can detect diffusional and spatial dynamics of juxtacrine receptors. Interaction dynamics of juxtacrine receptors is critical for the signaling, and hence we also develop such a technique. For example, MagAPPs (i.e., Magnetically Amplified Protein-Protein signals), a new nanotechnology tool, allows the investigators to measure cis-interactions of juxtacrine receptors precisely and quantitatively. Ultimately, we aim to establish platforms that can be applicable for diverse juxtacrine receptor signaling systematically and quantitatively.
Interrogate juxtacrine signaling using the cutting-edge tools
Using these new cutting-edge techniques, we address several long-standing questions in key juxtacrine signaling, including Notch (key cell-cell communication receptors), cadherins (mechanical communication), programmed death-1 (PD1, immunological checkpoint receptor) (e.g., CD80, PD-L1), neuroligin (synaptogenesis), and acetylcholine receptors (neuromuscular junction). Examples include (1) identifying Notch as a true-mechanoreceptor, (2) unraveling the biophysical mechanism regulating γ-secretase processing of Notch and amyloid precursor proteins (APPs), (3) revealing the differential roles of spatial and mechanical cues in cadherin-mediated adhesion junction formation, (4) determining anomalous and restricted diffusion of Notch due to size-based exclusion, and (5) measuring cis-interaction dynamics of PD-L1 quantitatively. Ultimately, using the technological platforms, we aim to present general biophysical principles regulating juxtacrine signaling.